- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
SPLICE VARIANTS (ISOFORMS)
One of the many versatile and flexible aspects of our genome's inner workings is the fact that a single gene can code for more than a single protein. Once a gene has been transcribed, the mRNA can be spliced in a variety of ways; introns are removed, but depending on the desired protein, certain exons are spliced out while others are left to be translated. A splice variant, or isoform, is one of the products of this process. The Genome Browser can be used to see all of a gene's different isoforms. Some genes have only one isoform while others have more than ten. The "UCSC Genes" track on the hg19 genome assembly is just one of the many tracks that shows isoform/splice variants.
A gene that has 15 different isoforms, SORBS1, is involved in insulin stimulation (Figure 1). SORBS1 is read on the negative strand, signified by the leftward facing arrows (denoting introns). With a quick look over, one can see some obvious differences among the splice variants; some isoforms have exons (denoted by boxes) that other isoforms do not, and start and stop sites vary widely. Once translated, these transcript differences produce different proteins which serve unique functions depending on the type of cell.
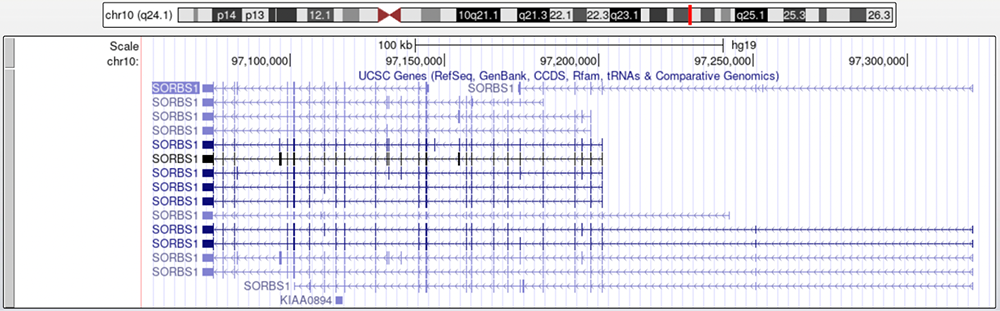
Figure 1.
SORBS1 on the "UCSC Genes" track. SORBS1 is read on the opposite, negative strand, hence the left-ward facing intron arrows. There are 15 isoforms shown on screen.
To understand the display conventions, including the difference in colors:
 colors.
colors.
 https://genome.ucsc.edu/s/education/hg19_SORBS1
https://genome.ucsc.edu/s/education/hg19_SORBS1
TISSUE-SPECIFIC ISOFORM EXPRESSION
Gene expression varies from cell-type to cell-type because different cells are going to need different proteins to function in their assigned way. A muscle cell is going to be made of different proteins and have different metabolic needs than a brain cell since they have different functions; this means genes, and therefore isoforms, are going to be expressed at different frequencies within each cell-type. This is where tissue-specific isoform expression comes into play.
GTEx is a database that has five tracks on the Genome Browser that can be used to visualize tissue-specific isoform expression. Found in the "Expression" section are the tracks "GTEx Gene V8", "GTEx Gene", and "GTEx Transcript". The two former tracks both average gene expression in cell types from all of the gene's isoforms, shown in Figure 2 ("V8" has an additional tissue type). However, "GTEx Transcript" shows cell-type gene expression for each individual isoform. Since the "GTEx Transcript" track isolates splice variant expression, let's choose two isoforms from SORBS1 and compare their tissue-specific gene expression.
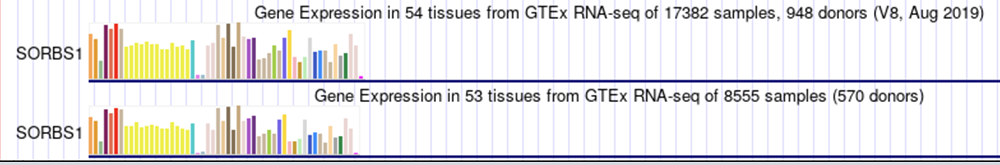
Figure 2.
"GTEx Gene V8" and "GTEx Gene" tracks on the SORBS1 gene.
Each colored bar represents a different cell type:
 cell types.
cell types.
 https://genome.ucsc.edu/s/education/hg19_SORBS1gtex
https://genome.ucsc.edu/s/education/hg19_SORBS1gtex
Figure 3a shows a custom track which isolates two isoforms of SORBS1, ENST00000371227.8_1 (dubbed ENST227 from this point for convenience) and ENST00000371249.6_1 (dubbed ENST249). ENST227 includes seven exons that ENST249 does not. This is visually clear as ENST249 is missing some exons that are present on ENST227. The mature transcripts encode proteins of 816 and 1266 amino acids respectively. As seen in Figure 3a, below the isoform custom track is the "GTEx Transcript" track, which shows two individual bar charts, for these isoforms, illustrating its expression frequencies in different tissue types. In the close-up, Figure 3b, three exons of one isoform can be seen that are absent from the other.

Figure 3a.
Top track is a custom track isolating two isoforms, ENST00000371227.8_1 and ENST00000371249.6_1 of the SORBS1 gene.
Below that is the "GTEx Transcript" track and each isoform�s corresponding tissue-specific expression graph.
 https://genome.ucsc.edu/s/education/hg19_SORBSct
https://genome.ucsc.edu/s/education/hg19_SORBSct

Figure 3b. Zoomed-in section of the SORBS1 gene, illustrating how ENST00000371227.8_1 contains exons missing from ENST00000371249.6_1.
There are lots of differences between these two transcript's levels of gene expression in varying tissue types; ENST227 has high frequencies of expression in muscle-skeletal and heart cells (two types), while ENST249 has high expression frequencies in colon, esophagus, bladder, artery, and fallopian tube cells. Insulin demands may be very different from tissue to tissue, and the genome allows for different isoforms of SORBS1 to be produced in order to address the needs of individual cell types. The data provided in the GTEx tracks are the raw material for scientists seeking to understand how gene expression differs in different cell and tissue types.
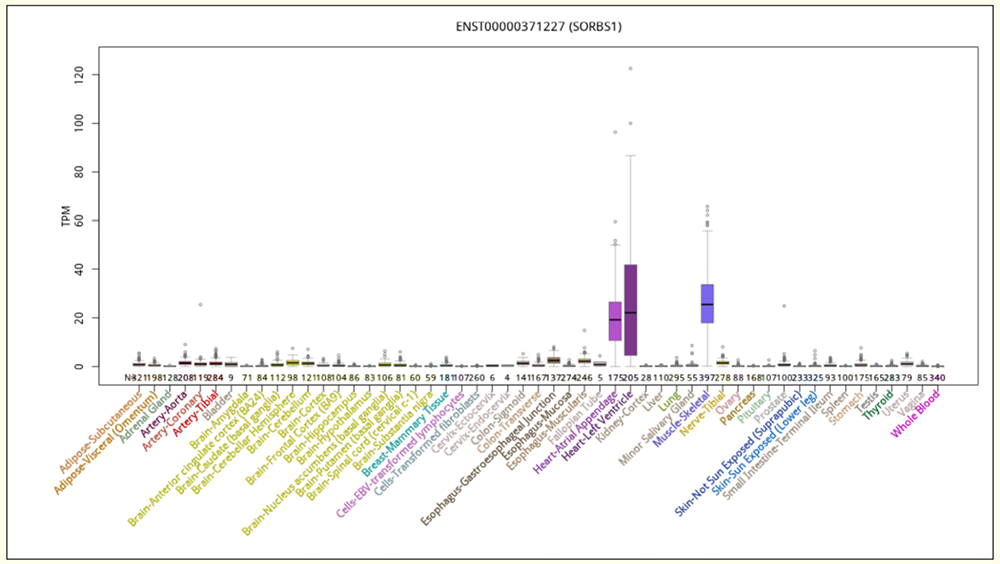
Figure 4. Expanded transcript expression in 53 tissue-types from GTEx for isoform ENST00000371227 (ENST227).
 GTEX details.
GTEX details.
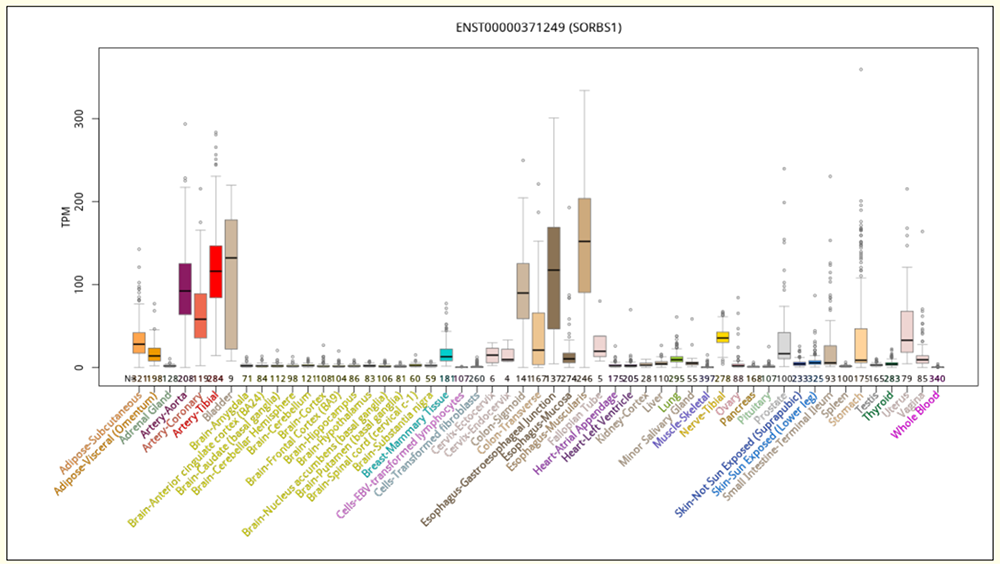
Figure 5. Expanded transcript expression in 53 tissue-types from GTEx for isoform ENST00000371249 (ENST249).
 GTEX details.
GTEX details.
Written by Zoë Shmidt, UCSC. Major: BA, Biological Anthropology