- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
EXPLORING CAG REPEATS IN HUNTINGTON'S DISEASE
CAG repeats are repeated sequences encoding anywhere from 6-37 glutamine amino acids. This particular sequence in Figure 1 below encodes the 5' end of the Huntington or HTT gene and shows a Genome Browser with perfect matches to an 18-base sequence consisting of six repeats of CAG (CAG6).
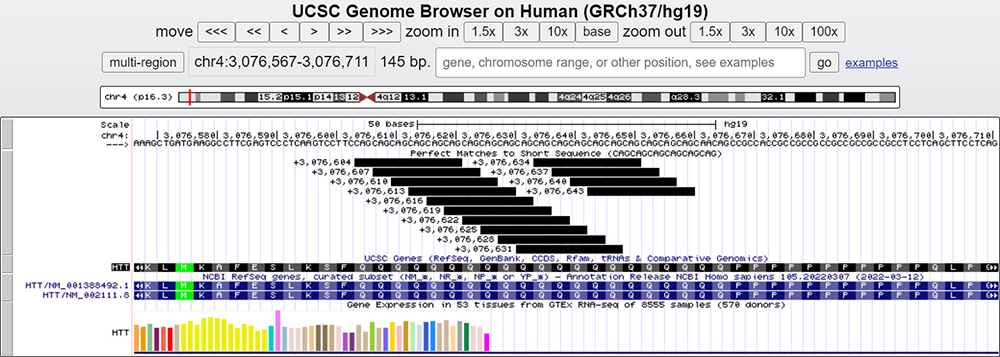
Figure 1. Main session window for the CAG repeats within the Huntington or HTT gene.
 https://genome.ucsc.edu/s/education/htt_main
https://genome.ucsc.edu/s/education/htt_main
In this session, we can see exactly 21 repeats of our CAG sequence in the hg19 human reference assembly. Something we can notice directly off our session window is the gene expression of HTT (Figure 2). If we click on the following:

Figure 2. Gene expression of HTT gene.
We are brought to the following display page (Figure 3):

Figure 3. Gene expression page for HTT gene with a more detailed general overview for expression of the gene.
In Figure 3, we can observe the varying expression of the HTT gene throughout the human body (Figure 2 shows the same data, but a log transform compresses the differences). Excluding Epstein-Barr virus-transformed cells (the pink peak), our first observation is that the expression of the HTT gene is high in regions of the brain (yellow). This can help explain the correlation between the HTT gene expression and Huntington's Disease which causes slow degeneration of brain tissue notably within the cerebellum. Furthermore, high gene expression of HTT within EBV-transformed lymphocytes could correlate with a hyperactive immune system with too many white blood cells — known to occur when patients have Huntington's Disease. We know that CAG repeats of more than 36 usually result in Huntington's Disease. As we can see, the cerebellum is marked as having the highest gene expression of the brain tissues on the graph in Figure 3.
Let's return to the main session (Figure 1) and take a look at another observation.
In Figure 1, we see the CAG repeats in the "Perfect Matches" track. In this particular window for this particular chromosome, there are 21 consecutive glutamine codons in the reference assembly. We know through studies that repeats within this gene of 26 glutamines or fewer are "normal" and will not result in Huntington's Disease. Repeats within the range of 27-35 fall within the "intermediate" category, where the patient is not at risk of developing symptoms of HD, but can conceive offspring within the HD-causing range. "Reduced Penetrance" are patients with 36-39 repeats. In this range, the patient may or may not go on to develop symptoms of HD, however, their children are at high risk of having HD. Finally, the last category, "Affected", applies to patients with 40 or more repeats. In this scenario, the patient will develop symptoms of HD throughout the course of their life. The more repeats present, the earlier the onset of symptoms. Currently, the exact reason why glutamine repeats cause this disease remains unknown. However, some scientists and studies speculate "instability" within the DNA structure as a possible cause.
In Figure 4, if we closely examine the nucleotide bases in our CAG repeat sequence, we can notice that amidst the CAG repeats, there is the insertion of one glutamine with the codons of CAA instead of CAG:

Figure 4. Zoom in on the CAG repeat sequence within the HTT gene, showing a particular section of the CAG nucleotide sequence. In the middle of the close-up, we can see a CAA codon.
Something interesting to note is that our CAG repeats subside almost immediately after this CAA. To look into this further, we can use the Browser to see if this is a common occurrence throughout the genome. In the next article, we will conduct a genome-wide search for other CAG and CAA repeats and look for genes that contain this motif to see if there's any correlation with genetic diseases/disorders.
Written by Mateo Etcheveste, UCSC. Major: BS, Biomolecular Engineering