- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
SPLICING IN THE GENOME BROWSER
Messenger RNA precursor, more commonly known as pre-mRNA contains both introns and exons. Through the process of splicing, these introns are removed, leaving behind the exons in a mature mRNA which then code for a functioning protein. The actual process of splicing itself is rather complex and intricate. If you are looking for an in-depth explanation, the following link can provide one: mRNA Splicing - Biology LibreTexts.
In today's Genome Browser example, we will be looking at splicing throughout the genome, and figuring out how to find spliced isoforms of a particular gene. Currently, there are multiple data tracks on the browser we can use to explore alternative splicing; however, we will only be using two today: "SIB Alt-Splicing" and "Spliced ESTs." For this example, we will be looking at the FGFR2 gene (Figure 1), described as follows: "The protein encoded by this gene is a member of the fibroblast growth factor receptor family, where amino acid sequence is highly conserved between members and throughout evolution." FGFR2 contains several spliced variants through a process known as alternative splicing. Alternative splicing is the selection of different combinations of splice sites within a pre-mRNA to produce variably spliced mRNAs. For this specific example, we will be zooming in on one particular region of the large FGFR2 gene with multiple splice variants in order to provide a clearer understanding.

Figure 1. FGFR2 on the "UCSC Genes" (green background)
and "NCBI Genes" tracks.
FGFR2 is read on the opposite, negative strand, hence the leftward facing intron arrows.
View a live Browser session:
 http://genome.ucsc.edu/s/education/fgfr2
http://genome.ucsc.edu/s/education/fgfr2
"Spliced ESTs" Data Track
The first data track we will look at is "Spliced ESTs." This data track shows alignments between human expressed sequence tags (ESTs) in GenBank and the genome. ESTs are single-read sequences, typically about 500 bases in length, that usually represent fragments of transcribed genes. Upon setting this track to pack, the browser will load the view seen in Figure 2.
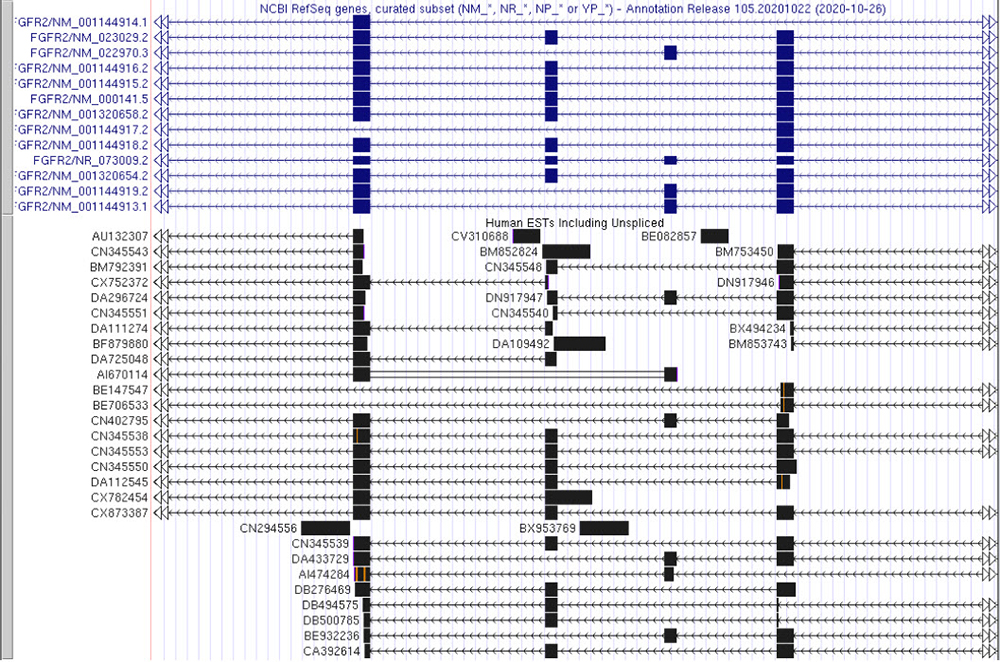
Figure 2. "Spliced ESTsi" data track set to pack showing varying gene arrangements for the FGFR2 gene. Each EST is labeled with the corresponding tag to the left of its sequence.
The "Spliced ESTsi" track allows us to analyze sequences of FGFR2 gene with different exon arrangements.
The black boxes in the data track represent reads that match the "UCSC Genes" and/or
"NCBI Genes"/GenBank database. Orange/purple lines match an unalignable query sequence, orange for the middle of a sequence, and purple for the beginning or end.
And green signifies a poly-A tail or poly-T head that does not align to the genome (more details listed here:
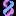 "Spliced ESTs").
Comparing the various arrangements we can observe reads with spliced-out exons in several different combinations.
"Spliced ESTs").
Comparing the various arrangements we can observe reads with spliced-out exons in several different combinations.
Indicated by the red arrow in Figure 3 is our first example of an exon spliced out of an EST sequence read: CN402795. The yellow highlight marks exon 6 of the FGFR2 gene, which was spliced out of EST CN402795. CN402795 represents a splice variant of the FGFR2 gene sequence that lacks exon 6.
Furthermore, in Figure 3, the orange highlight marks exon 7,where we can see within the purple circle two more examples of EST reads with exons spliced out: BE706533 and BE147547. These two variants are missing exon 7 (orange highlight) and are also missing exon 6 (yellow highlight).
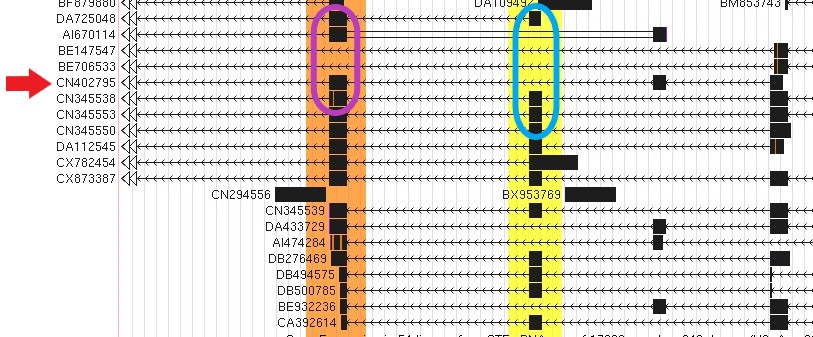
Figure 3.
 http://genome.ucsc.edu/s/education/fgfr2_highlights.
CN402795 sequence read (red arrow) missing exon 6 for the FGFR2 gene (found within the yellow highlight in the blue circle). In the orange highlighted region, we can see two more splice variants BE706533 and BE147547 missing exon 7.
http://genome.ucsc.edu/s/education/fgfr2_highlights.
CN402795 sequence read (red arrow) missing exon 6 for the FGFR2 gene (found within the yellow highlight in the blue circle). In the orange highlighted region, we can see two more splice variants BE706533 and BE147547 missing exon 7.
"SIB Alt-Splicing" Data Track
The next splicing data track "SIB Alt-Splicing", shows graphs constructed by analyzing experimental RNA transcripts and serves as the basis for the predicted alternative splicing transcripts shown in the SIB Genes track. In Figure 4 we can observe the "SIB Alt-Splicing" data track set to full for the FGFR2 gene. Here, the lines mark numerous possible spliced variants from registered RNA transcripts. Each line represents a recorded RNA splice event supported by a transcript and the exons it includes while a skipped exon implies that the exon was spliced out and not recorded in the transcript. Notice how in Figure 4, two separate colored lines include different exons within their path — representing the different spliced variant isoforms. The red line includes three exons while the purple line has skipped the first two exons that are included in the transcript in red.
Figure 4. "SIB Alt-Splicing" data track showing part of the FGFR2 gene.
Red
lines have been added to represent the splice pattern of the EST indicated by the red arrow in Figure 3 (CN402795).
Purple lines have been added to represent the splice variant isoforms
for the FGFR2 gene indicated by the purple oval in Figure 3 (BE706533 and BE147547).
For more information about this data track, see the
 "SIB Alt-Splicing" Display Conventions Page
"SIB Alt-Splicing" Display Conventions Page
Clicking on any of the block boxes within the data track will take you to the "SIB Alt-Splicing" details page for this gene. In this case, clicking on it will bring you to the page seen in Figure 5 for FGFR2.
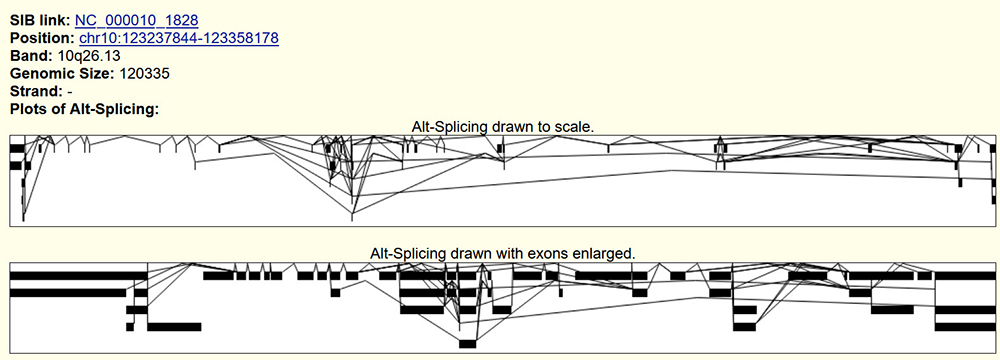
Figure 5. "SIB Alt-Splicing" data track main page for the FGFR2 gene. Here, a more detailed representation of the possible RNA transcripts with their unique spliced variants is included. Link to example page within the FGFR2 gene:
 Example 1
Example 1
Something worth noting is that these graphs show the variation throughout the entire gene, not the particular section we zoomed in on for FGFR2 above. This explains why more exons are included in the pictures and the significant increase in variation among the individual transcripts. In Figure 5, the splicing variation is much more apparent throughout the entirety of the gene as we can see certain lines touching only specific exons while others skip over them.
Written by Mateo Etcheveste, UCSC. Major: BS, Biomolecular Engineering