- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
FOXP2: SPEECH DEVELOPMENT & EVOLUTION
The FOXP2 gene is associated with the developmental processes of language and speech in humans. It is also thought to be a part of why humans are capable of speech while chimpanzees and other apes are not. The Genome Browser can be utilized to visualize the pathogenic components and evolutionary changes of FOXP2.
THE ":KE FAMILY"
FOXP2 was found to be associated with language through a British family dubbed the "KE Family" or "KE pedigree." Almost half of the members for the past three generations have developmental verbal dyspraxia, a speech disorder that is characterized by difficulty understanding grammatical conventions, a limited vocabulary, stuttering, and restricted pronunciation abilities. Members of the KE family also suffered from deformed orofacial morphology and "abnormal development of a number of cortical and subcortical areas" (Watkins 2011). After multiple investigations, neuroscientists, linguists, and doctors found the KE family's disease to be of genetic origin, specifically a mutation within the FOXP2 gene. The chromosomal region that FOXP2 lies within was dubbed SPCH1, as the 70 genes within this locus are almost all related to speech development and cognition.
Figure 1 shows the nucleotide change found within the affected members of the KE family pedigree. The green box on the "OMIM Allelic Variant Phenotypes" track marks the variant's position within FOXP2. The isoform of interest has its 553rd amino acid (as reported in Watkins) marked by the yellow highlighted column (R 553, fourth isoform from the top). The variation is a G to A change which results in a different amino acid than the one encoded by the reference genome; arginine (R) to histidine (H).
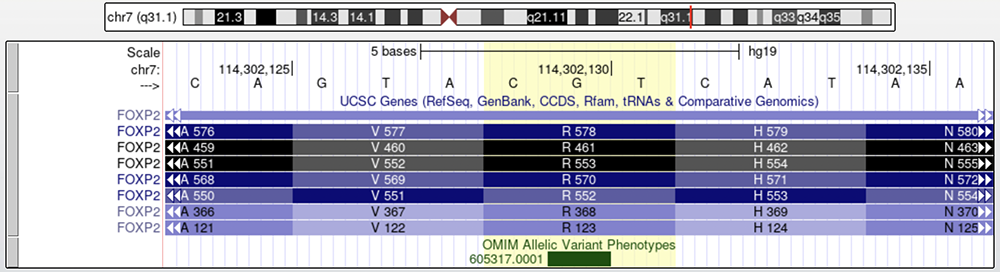
Figure 1.
(
 http://genome.ucsc.edu/s/education/hg19_FOXP2p.R553H).
FOXP2 on the "UCSC Genes Track." The "OMIM Allelic Variant Phenotypes" track marks the variant found
within the KE pedigree [p.(R553H), exon 14]; the isoform of interest has the amino acid "R553"
within the highlighted column. The highlighted column shows the entire codon affected by the nucleotide change.
http://genome.ucsc.edu/s/education/hg19_FOXP2p.R553H).
FOXP2 on the "UCSC Genes Track." The "OMIM Allelic Variant Phenotypes" track marks the variant found
within the KE pedigree [p.(R553H), exon 14]; the isoform of interest has the amino acid "R553"
within the highlighted column. The highlighted column shows the entire codon affected by the nucleotide change.
Clicking on the green "OMIM Variant" box will bring you to the variants detail page (Figure 2). Navigating to the link found at the top of the details page will take you to OMIM's database and a further description of the variant and its phenotypic consequences. On OMIM's website, the KE family pedigree is mentioned as the original report of this variant. If interested in more information regarding the pathogenic effects of this variant, one could also enable the ClinVar or other medical tracks on the genome browser.
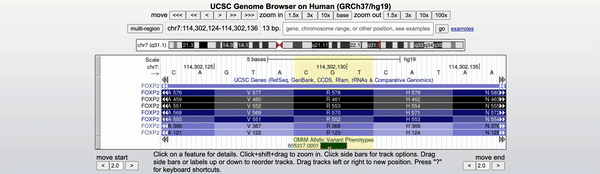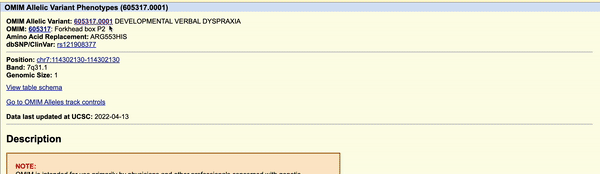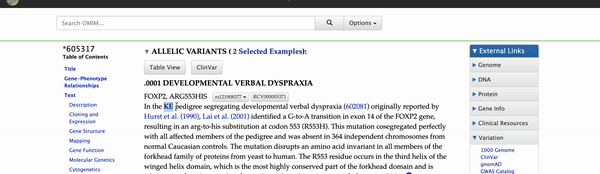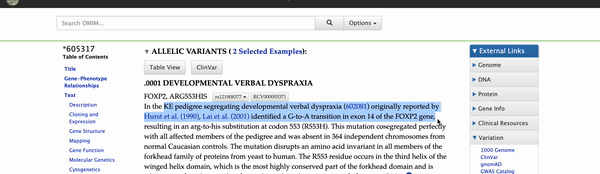
Figure 2. Video illustrating how to navigate to the OMIM database on the 605317.0001 variant
EVOLUTIONARY CONSERVATION AND CHANGES
Most of FOXP2 is heavily conserved between humans and other mammals. However, there are two amino acid differences between the genomes of humans and other primates that have been of interest lately, as they are thought to be connected to why humans are capable of speech while chimpanzees and other apes are not. Figures 3 and 4 isolate these two amino acid changes on the Genome Browser.
Figure 3 illustrates the first of the two amino acid changes associated with development of speech in humans. The "UCSC Genes" track shows multiple isoforms, two of which have an asparagine (N) at position 303 on exon 7 (as described in Zhang et al., 2002) marked by the yellow highlight. Primates instead possess a threonine (T).
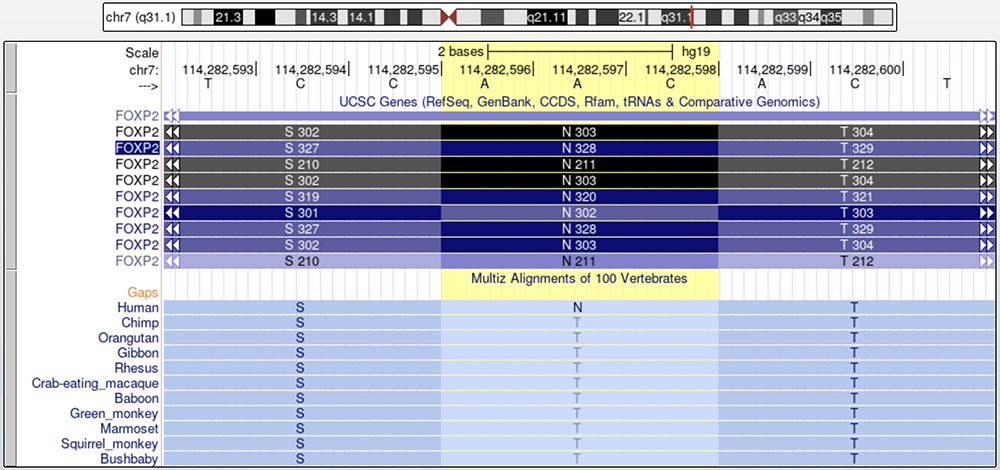
Figure 3.
(
 http://genome.ucsc.edu/s/education/hg19_FOXP2fig3).
The highlighted column isolates the first of the amino acid changes between humans and other primates thought to be associated with the development of speech.
http://genome.ucsc.edu/s/education/hg19_FOXP2fig3).
The highlighted column isolates the first of the amino acid changes between humans and other primates thought to be associated with the development of speech.
Figure 4 illustrates the second of the two amino acid changes that are associated with language ability in humans and inability in other primates using a highlighted column. There are two isoforms that meet this criterion and have a serine (S) in the 325 position (Zhang et al., 2002): The second and fifth isoforms in Figure 4. The "Multiz Alignments of 100 Vertebrates" track shows the corresponding amino acids of the reference sequences of humans and a variety of other primate species. While humans have an S in this position, or a serine, all other primates possess a N, or an asparagine.
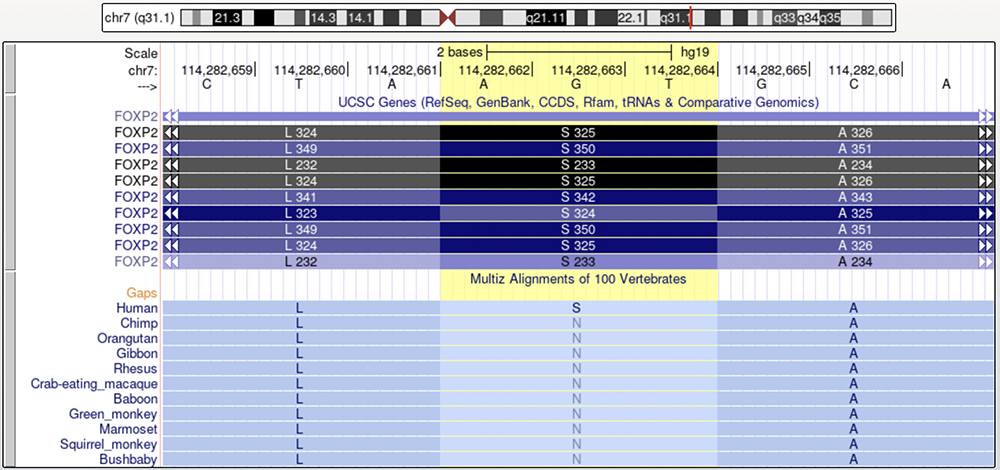
Figure 4.
(
 http://genome.ucsc.edu/s/education/hg19_FOXP2fig4).
The highlighted column isolates the second of the amino acid changes between humans and other primates thought to be associated with the development of speech.
http://genome.ucsc.edu/s/education/hg19_FOXP2fig4).
The highlighted column isolates the second of the amino acid changes between humans and other primates thought to be associated with the development of speech.
The FOXP2 gene is essential in our cognitive ability to understand language and developing proficient speech; The KE family's private variant within FOXP2 resulting in developmental verbal dyspraxia supports this. Comparing the human reference genome seen on the Genome Browser to other primates' sequences of FOXP2 also sheds light on the association between FOXP2 and language. The two amino acid differences seen in Figure 3 and Figure 4 illustrate the genotypical evolutionary changes that we think contributed to hominin development and comprehension of language. These variants may have been naturally selected for in early genus Homo populations, as language became increasingly beneficial for co-operation, and therefore survival, in dangerous or severe environments.
REFERENCES
Watkins, 2011. KE Family: Gene Expression to Neurobiology and Behavior
Zhang et al., 2002. Accelerated Protein Evolution and Origins of Human-Specific Features: FOXP2
Evolution of a Single Gene Linked to Language
Human-specific transcriptional regulation of CNS development genes by FOXP2
Written by Zoë Shmidt, UCSC. Major: BA, Biological Anthropology